核密度估计¶
核密度估计是使用核函数 \(K(u)\) 估计未知概率密度函数的过程。直方图统计在某些任意区域中的数据点的数量,而核密度估计是一个定义为每个数据点上的核函数之和的函数。核函数通常具有以下属性
对称性,使得 \(K(u) = K(-u)\)。
归一化,使得 \(\int_{-\infty}^{\infty} K(u) \ du = 1\) 。
单调递减,使得 \(K'(u) < 0\) 当 \(u > 0\) 时。
期望值为零,使得 \(\mathrm{E}[K] = 0\)。
有关核密度估计的更多信息,请参阅例如 维基百科 - 核密度估计。
单变量核密度估计器在 sm.nonparametric.KDEUnivariate 中实现。在本示例中,我们将展示以下内容
基本用法,如何拟合估计器。
使用
bw参数改变核带宽的效果。使用
kernel参数可用的各种核函数。
[1]:
%matplotlib inline
import numpy as np
from scipy import stats
import statsmodels.api as sm
import matplotlib.pyplot as plt
from statsmodels.distributions.mixture_rvs import mixture_rvs
单变量示例¶
[2]:
np.random.seed(12345) # Seed the random number generator for reproducible results
我们创建一个双峰分布:两个均值分别位于 -1 和 1 的正态分布的混合。
[3]:
# Location, scale and weight for the two distributions
dist1_loc, dist1_scale, weight1 = -1, 0.5, 0.25
dist2_loc, dist2_scale, weight2 = 1, 0.5, 0.75
# Sample from a mixture of distributions
obs_dist = mixture_rvs(
prob=[weight1, weight2],
size=250,
dist=[stats.norm, stats.norm],
kwargs=(
dict(loc=dist1_loc, scale=dist1_scale),
dict(loc=dist2_loc, scale=dist2_scale),
),
)
最简单的非参数密度估计技术是直方图。
[4]:
fig = plt.figure(figsize=(12, 5))
ax = fig.add_subplot(111)
# Scatter plot of data samples and histogram
ax.scatter(
obs_dist,
np.abs(np.random.randn(obs_dist.size)),
zorder=15,
color="red",
marker="x",
alpha=0.5,
label="Samples",
)
lines = ax.hist(obs_dist, bins=20, edgecolor="k", label="Histogram")
ax.legend(loc="best")
ax.grid(True, zorder=-5)
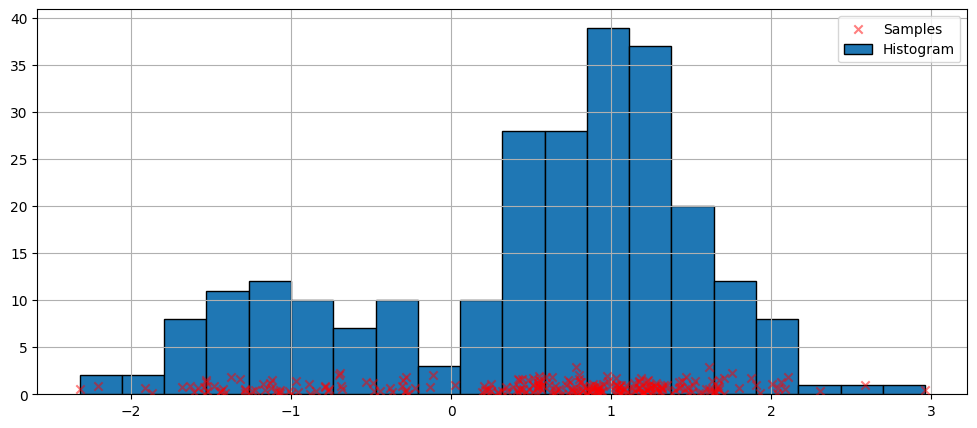
使用默认参数拟合¶
上面的直方图是不连续的。为了计算连续的概率密度函数,我们可以使用核密度估计。
我们使用 KDEUnivariate 初始化单变量核密度估计器。
[5]:
kde = sm.nonparametric.KDEUnivariate(obs_dist)
kde.fit() # Estimate the densities
[5]:
<statsmodels.nonparametric.kde.KDEUnivariate at 0x7fed558239a0>
我们展示了拟合的图形以及真实分布。
[6]:
fig = plt.figure(figsize=(12, 5))
ax = fig.add_subplot(111)
# Plot the histogram
ax.hist(
obs_dist,
bins=20,
density=True,
label="Histogram from samples",
zorder=5,
edgecolor="k",
alpha=0.5,
)
# Plot the KDE as fitted using the default arguments
ax.plot(kde.support, kde.density, lw=3, label="KDE from samples", zorder=10)
# Plot the true distribution
true_values = (
stats.norm.pdf(loc=dist1_loc, scale=dist1_scale, x=kde.support) * weight1
+ stats.norm.pdf(loc=dist2_loc, scale=dist2_scale, x=kde.support) * weight2
)
ax.plot(kde.support, true_values, lw=3, label="True distribution", zorder=15)
# Plot the samples
ax.scatter(
obs_dist,
np.abs(np.random.randn(obs_dist.size)) / 40,
marker="x",
color="red",
zorder=20,
label="Samples",
alpha=0.5,
)
ax.legend(loc="best")
ax.grid(True, zorder=-5)
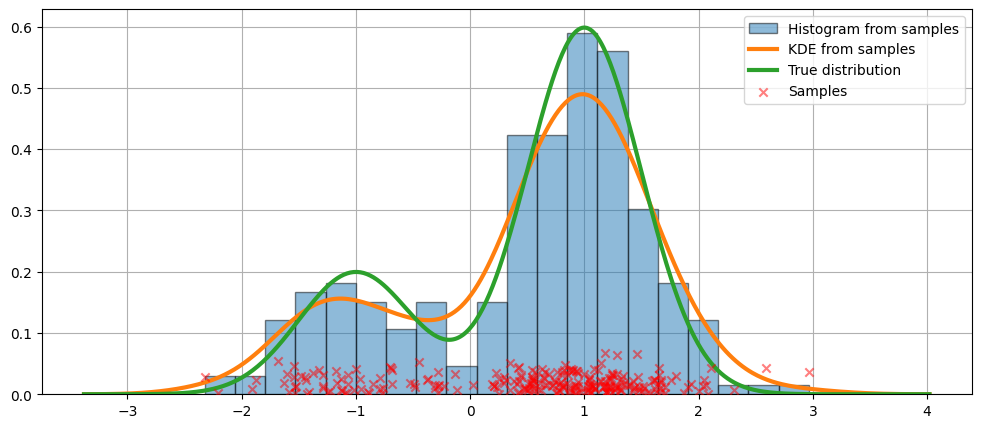
在上面的代码中,使用了默认参数。我们也可以改变核的带宽,正如我们现在将看到的那样。
使用 bw 参数改变带宽¶
可以使用 bw 参数调整核的带宽。在以下示例中,带宽 bw=0.2 似乎可以很好地拟合数据。
[7]:
fig = plt.figure(figsize=(12, 5))
ax = fig.add_subplot(111)
# Plot the histogram
ax.hist(
obs_dist,
bins=25,
label="Histogram from samples",
zorder=5,
edgecolor="k",
density=True,
alpha=0.5,
)
# Plot the KDE for various bandwidths
for bandwidth in [0.1, 0.2, 0.4]:
kde.fit(bw=bandwidth) # Estimate the densities
ax.plot(
kde.support,
kde.density,
"--",
lw=2,
color="k",
zorder=10,
label="KDE from samples, bw = {}".format(round(bandwidth, 2)),
)
# Plot the true distribution
ax.plot(kde.support, true_values, lw=3, label="True distribution", zorder=15)
# Plot the samples
ax.scatter(
obs_dist,
np.abs(np.random.randn(obs_dist.size)) / 50,
marker="x",
color="red",
zorder=20,
label="Data samples",
alpha=0.5,
)
ax.legend(loc="best")
ax.set_xlim([-3, 3])
ax.grid(True, zorder=-5)
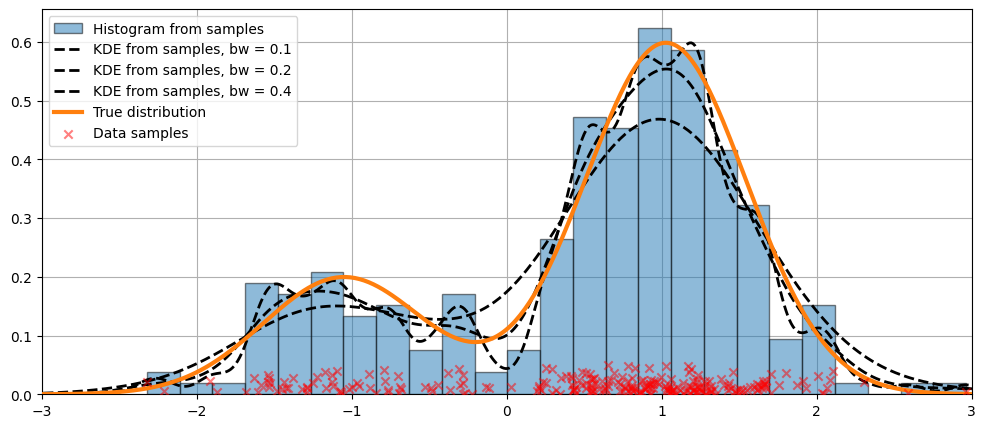
比较核函数¶
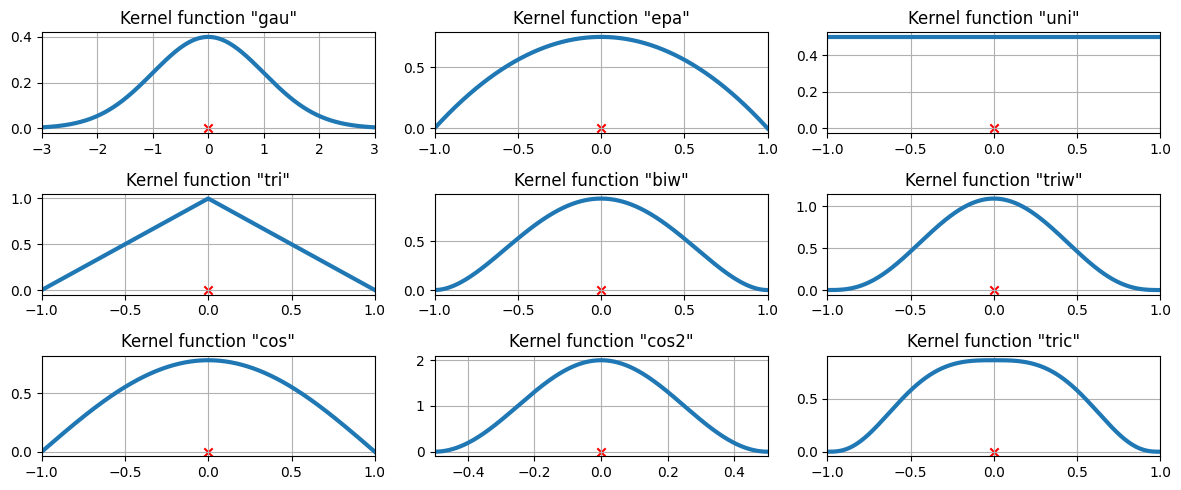
在上面的示例中,使用了高斯核。还提供了一些其他核。
[8]:
from statsmodels.nonparametric.kde import kernel_switch
list(kernel_switch.keys())
[8]:
['gau', 'epa', 'uni', 'tri', 'biw', 'triw', 'cos', 'cos2', 'tric']
可用的核函数¶
[9]:
# Create a figure
fig = plt.figure(figsize=(12, 5))
# Enumerate every option for the kernel
for i, (ker_name, ker_class) in enumerate(kernel_switch.items()):
# Initialize the kernel object
kernel = ker_class()
# Sample from the domain
domain = kernel.domain or [-3, 3]
x_vals = np.linspace(*domain, num=2 ** 10)
y_vals = kernel(x_vals)
# Create a subplot, set the title
ax = fig.add_subplot(3, 3, i + 1)
ax.set_title('Kernel function "{}"'.format(ker_name))
ax.plot(x_vals, y_vals, lw=3, label="{}".format(ker_name))
ax.scatter([0], [0], marker="x", color="red")
plt.grid(True, zorder=-5)
ax.set_xlim(domain)
plt.tight_layout()
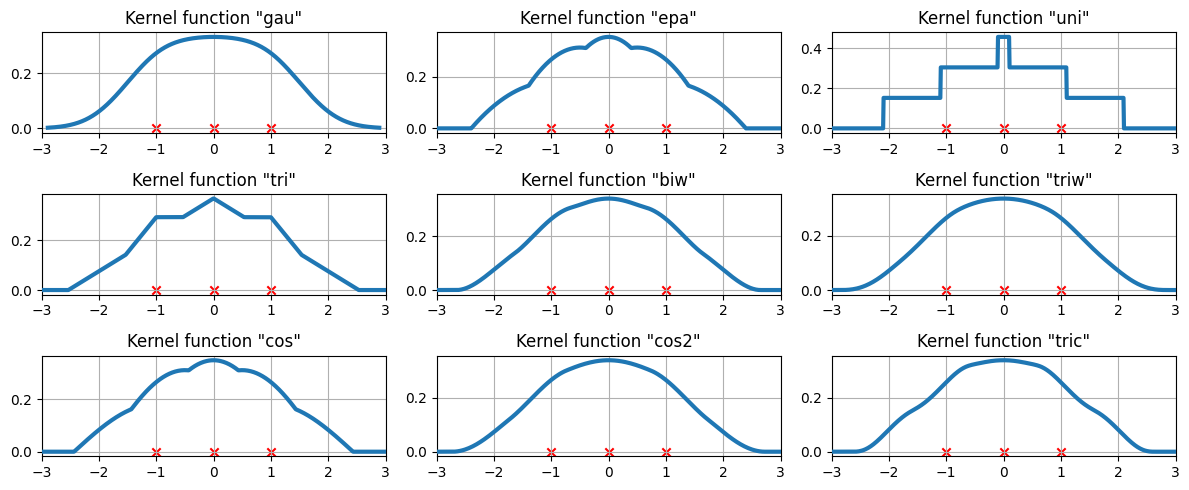
在三个数据点上的可用核函数¶
我们现在将研究核密度估计将如何拟合三个等间距的数据点。
[10]:
# Create three equidistant points
data = np.linspace(-1, 1, 3)
kde = sm.nonparametric.KDEUnivariate(data)
# Create a figure
fig = plt.figure(figsize=(12, 5))
# Enumerate every option for the kernel
for i, kernel in enumerate(kernel_switch.keys()):
# Create a subplot, set the title
ax = fig.add_subplot(3, 3, i + 1)
ax.set_title('Kernel function "{}"'.format(kernel))
# Fit the model (estimate densities)
kde.fit(kernel=kernel, fft=False, gridsize=2 ** 10)
# Create the plot
ax.plot(kde.support, kde.density, lw=3, label="KDE from samples", zorder=10)
ax.scatter(data, np.zeros_like(data), marker="x", color="red")
plt.grid(True, zorder=-5)
ax.set_xlim([-3, 3])
plt.tight_layout()
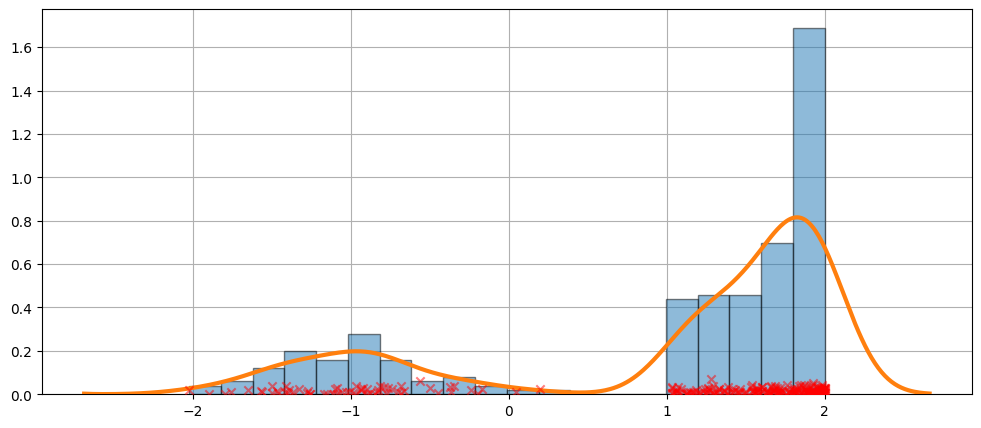
更难的情况¶
拟合并不总是完美的。请参见以下示例以了解更难的情况。
[11]:
obs_dist = mixture_rvs(
[0.25, 0.75],
size=250,
dist=[stats.norm, stats.beta],
kwargs=(dict(loc=-1, scale=0.5), dict(loc=1, scale=1, args=(1, 0.5))),
)
[12]:
kde = sm.nonparametric.KDEUnivariate(obs_dist)
kde.fit()
[12]:
<statsmodels.nonparametric.kde.KDEUnivariate at 0x7fed52347940>
[13]:
fig = plt.figure(figsize=(12, 5))
ax = fig.add_subplot(111)
ax.hist(obs_dist, bins=20, density=True, edgecolor="k", zorder=4, alpha=0.5)
ax.plot(kde.support, kde.density, lw=3, zorder=7)
# Plot the samples
ax.scatter(
obs_dist,
np.abs(np.random.randn(obs_dist.size)) / 50,
marker="x",
color="red",
zorder=20,
label="Data samples",
alpha=0.5,
)
ax.grid(True, zorder=-5)
KDE 是一个分布¶
由于 KDE 是一个分布,因此我们可以访问诸如以下属性和方法
熵评估cdficdfsfcumhazard
[14]:
obs_dist = mixture_rvs(
[0.25, 0.75],
size=1000,
dist=[stats.norm, stats.norm],
kwargs=(dict(loc=-1, scale=0.5), dict(loc=1, scale=0.5)),
)
kde = sm.nonparametric.KDEUnivariate(obs_dist)
kde.fit(gridsize=2 ** 10)
[14]:
<statsmodels.nonparametric.kde.KDEUnivariate at 0x7fed535c1c00>
[15]:
kde.entropy
[15]:
1.314324140492138
[16]:
kde.evaluate(-1)
[16]:
array([0.18085886])
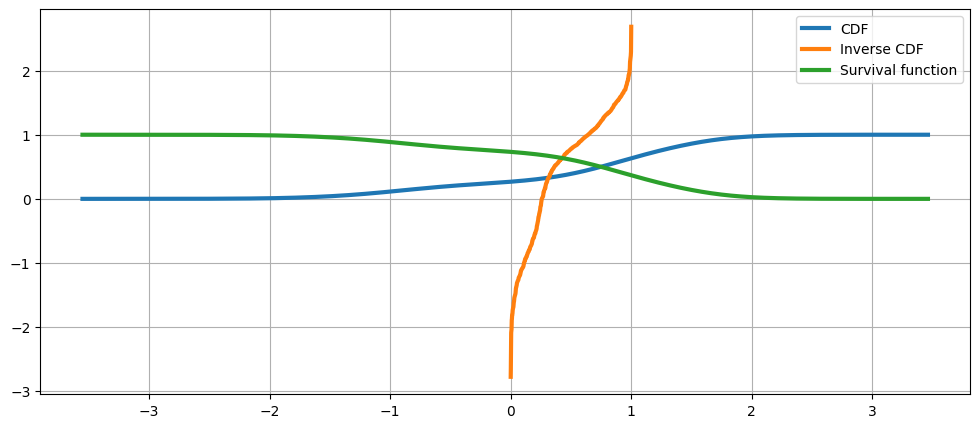
累积分布、其逆和生存函数¶
[17]:
fig = plt.figure(figsize=(12, 5))
ax = fig.add_subplot(111)
ax.plot(kde.support, kde.cdf, lw=3, label="CDF")
ax.plot(np.linspace(0, 1, num=kde.icdf.size), kde.icdf, lw=3, label="Inverse CDF")
ax.plot(kde.support, kde.sf, lw=3, label="Survival function")
ax.legend(loc="best")
ax.grid(True, zorder=-5)
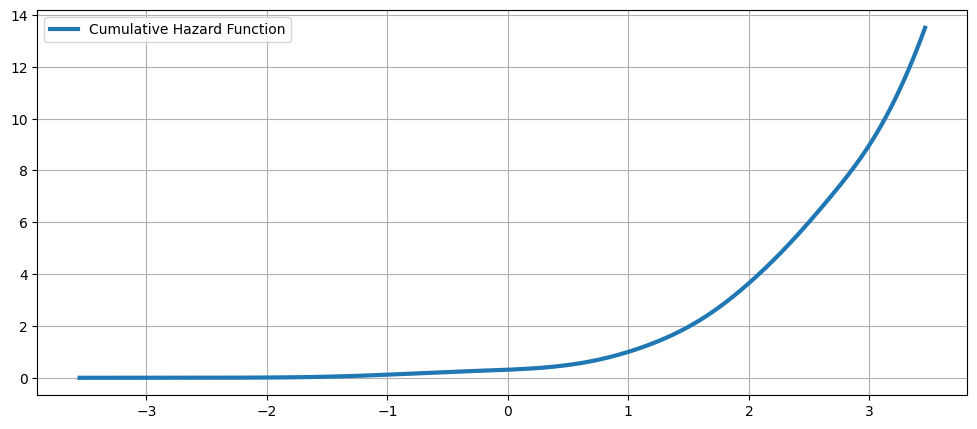
累积风险函数¶
[18]:
fig = plt.figure(figsize=(12, 5))
ax = fig.add_subplot(111)
ax.plot(kde.support, kde.cumhazard, lw=3, label="Cumulative Hazard Function")
ax.legend(loc="best")
ax.grid(True, zorder=-5)
上次更新:2024 年 10 月 3 日