statsmodels 中的元分析¶
Statsmodels 包含元分析的基本方法。本笔记本展示了当前用法。
状态:结果已针对 R meta 和 metafor 包进行验证。但是,API 仍处于实验阶段,将会发生变化。R meta 和 metafor 中可用的某些其他方法选项缺失。
对元分析的支持分为 3 个部分
效应量函数:目前包括
effectsize_smd计算标准化均值差的效应量及其标准误,effectsize_2proportions计算使用风险差、(对数)风险比、(对数)优势比或反正弦平方根变换比较两个独立比例的效应量The
combine_effects计算总体均值或效应的固定效应和随机效应估计。返回的结果实例包括一个森林图函数。辅助函数来估计随机效应方差,tau 平方
The estimate of the overall effect size in combine_effects can also be performed using WLS or GLM with var_weights.
最后,元分析函数目前不包括 Mantel-Hanszel 方法。但是,可以使用 StratifiedTable 直接计算固定效应结果,如下所示。
[1]:
%matplotlib inline
[2]:
import numpy as np
import pandas as pd
from scipy import stats, optimize
from statsmodels.regression.linear_model import WLS
from statsmodels.genmod.generalized_linear_model import GLM
from statsmodels.stats.meta_analysis import (
effectsize_smd,
effectsize_2proportions,
combine_effects,
_fit_tau_iterative,
_fit_tau_mm,
_fit_tau_iter_mm,
)
# increase line length for pandas
pd.set_option("display.width", 100)
示例¶
[3]:
data = [
["Carroll", 94, 22, 60, 92, 20, 60],
["Grant", 98, 21, 65, 92, 22, 65],
["Peck", 98, 28, 40, 88, 26, 40],
["Donat", 94, 19, 200, 82, 17, 200],
["Stewart", 98, 21, 50, 88, 22, 45],
["Young", 96, 21, 85, 92, 22, 85],
]
colnames = ["study", "mean_t", "sd_t", "n_t", "mean_c", "sd_c", "n_c"]
rownames = [i[0] for i in data]
dframe1 = pd.DataFrame(data, columns=colnames)
rownames
[3]:
['Carroll', 'Grant', 'Peck', 'Donat', 'Stewart', 'Young']
[4]:
mean2, sd2, nobs2, mean1, sd1, nobs1 = np.asarray(
dframe1[["mean_t", "sd_t", "n_t", "mean_c", "sd_c", "n_c"]]
).T
rownames = dframe1["study"]
rownames.tolist()
[4]:
['Carroll', 'Grant', 'Peck', 'Donat', 'Stewart', 'Young']
[5]:
np.array(nobs1 + nobs2)
[5]:
array([120, 130, 80, 400, 95, 170])
估计效应量标准化均值差¶
[6]:
eff, var_eff = effectsize_smd(mean2, sd2, nobs2, mean1, sd1, nobs1)
使用一步 chi2,DerSimonian-Laird 估计随机效应方差 tau¶
随机效应的 method 选项 method_re="chi2" 或 method_re="dl",这两个名称都接受。这通常被称为 DerSimonian-Laird 方法,它基于固定效应估计中基于 pearson chi2 的矩估计。
[7]:
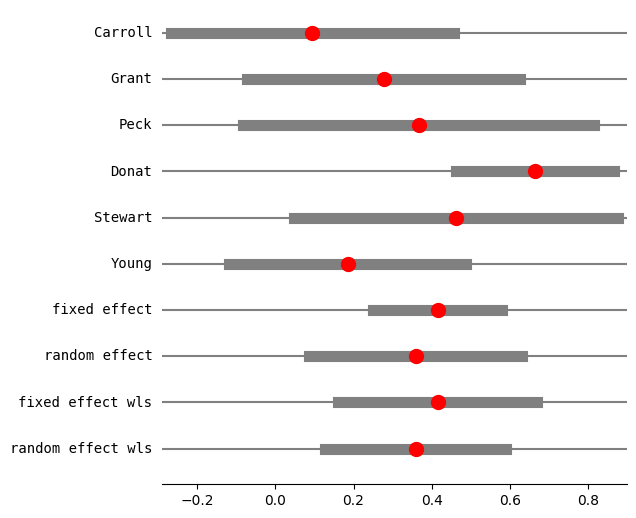
res3 = combine_effects(eff, var_eff, method_re="chi2", use_t=True, row_names=rownames)
# TODO: we still need better information about conf_int of individual samples
# We don't have enough information in the model for individual confidence intervals
# if those are not based on normal distribution.
res3.conf_int_samples(nobs=np.array(nobs1 + nobs2))
print(res3.summary_frame())
eff sd_eff ci_low ci_upp w_fe w_re
Carroll 0.094524 0.182680 -0.267199 0.456248 0.123885 0.157529
Grant 0.277356 0.176279 -0.071416 0.626129 0.133045 0.162828
Peck 0.366546 0.225573 -0.082446 0.815538 0.081250 0.126223
Donat 0.664385 0.102748 0.462389 0.866381 0.391606 0.232734
Stewart 0.461808 0.208310 0.048203 0.875413 0.095275 0.137949
Young 0.185165 0.153729 -0.118312 0.488641 0.174939 0.182736
fixed effect 0.414961 0.064298 0.249677 0.580245 1.000000 NaN
random effect 0.358486 0.105462 0.087388 0.629583 NaN 1.000000
fixed effect wls 0.414961 0.099237 0.159864 0.670058 1.000000 NaN
random effect wls 0.358486 0.090328 0.126290 0.590682 NaN 1.000000
[8]:
res3.cache_ci
[8]:
{(0.05,
True): (array([-0.26719942, -0.07141628, -0.08244568, 0.46238908, 0.04820269,
-0.1183121 ]), array([0.45624817, 0.62612908, 0.81553838, 0.86638112, 0.87541326,
0.48864139]))}
[9]:
res3.method_re
[9]:
'chi2'
[10]:
fig = res3.plot_forest()
fig.set_figheight(6)
fig.set_figwidth(6)
[11]:
res3 = combine_effects(eff, var_eff, method_re="chi2", use_t=False, row_names=rownames)
# TODO: we still need better information about conf_int of individual samples
# We don't have enough information in the model for individual confidence intervals
# if those are not based on normal distribution.
res3.conf_int_samples(nobs=np.array(nobs1 + nobs2))
print(res3.summary_frame())
eff sd_eff ci_low ci_upp w_fe w_re
Carroll 0.094524 0.182680 -0.263521 0.452570 0.123885 0.157529
Grant 0.277356 0.176279 -0.068144 0.622857 0.133045 0.162828
Peck 0.366546 0.225573 -0.075569 0.808662 0.081250 0.126223
Donat 0.664385 0.102748 0.463002 0.865768 0.391606 0.232734
Stewart 0.461808 0.208310 0.053527 0.870089 0.095275 0.137949
Young 0.185165 0.153729 -0.116139 0.486468 0.174939 0.182736
fixed effect 0.414961 0.064298 0.288939 0.540984 1.000000 NaN
random effect 0.358486 0.105462 0.151785 0.565187 NaN 1.000000
fixed effect wls 0.414961 0.099237 0.220460 0.609462 1.000000 NaN
random effect wls 0.358486 0.090328 0.181446 0.535526 NaN 1.000000
使用迭代的 Paule-Mandel 估计随机效应方差 tau¶
通常称为 Paule-Mandel 估计的方法是随机效应方差的矩估计方法,它在均值和方差估计之间迭代,直到收敛。
[12]:
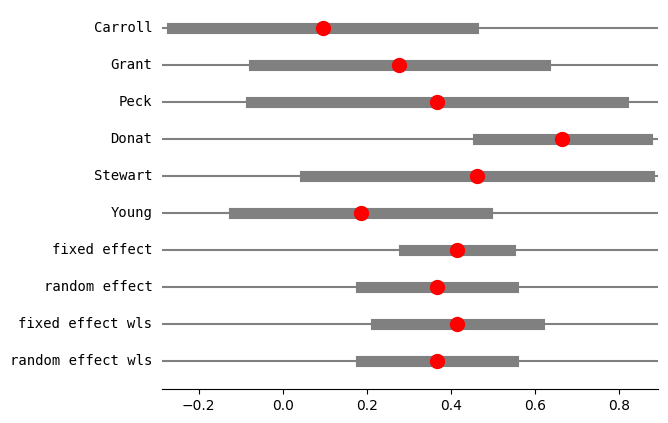
res4 = combine_effects(
eff, var_eff, method_re="iterated", use_t=False, row_names=rownames
)
res4_df = res4.summary_frame()
print("method RE:", res4.method_re)
print(res4.summary_frame())
fig = res4.plot_forest()
method RE: iterated
eff sd_eff ci_low ci_upp w_fe w_re
Carroll 0.094524 0.182680 -0.263521 0.452570 0.123885 0.152619
Grant 0.277356 0.176279 -0.068144 0.622857 0.133045 0.159157
Peck 0.366546 0.225573 -0.075569 0.808662 0.081250 0.116228
Donat 0.664385 0.102748 0.463002 0.865768 0.391606 0.257767
Stewart 0.461808 0.208310 0.053527 0.870089 0.095275 0.129428
Young 0.185165 0.153729 -0.116139 0.486468 0.174939 0.184799
fixed effect 0.414961 0.064298 0.288939 0.540984 1.000000 NaN
random effect 0.366419 0.092390 0.185338 0.547500 NaN 1.000000
fixed effect wls 0.414961 0.099237 0.220460 0.609462 1.000000 NaN
random effect wls 0.366419 0.092390 0.185338 0.547500 NaN 1.000000
[ ]:
示例 Kacker 实验室间平均值¶
在本例中,效应量是实验室中测量的平均值。我们将来自多个实验室的估计值结合起来,以估计总体平均值。
[13]:
eff = np.array([61.00, 61.40, 62.21, 62.30, 62.34, 62.60, 62.70, 62.84, 65.90])
var_eff = np.array(
[0.2025, 1.2100, 0.0900, 0.2025, 0.3844, 0.5625, 0.0676, 0.0225, 1.8225]
)
rownames = ["PTB", "NMi", "NIMC", "KRISS", "LGC", "NRC", "IRMM", "NIST", "LNE"]
[14]:
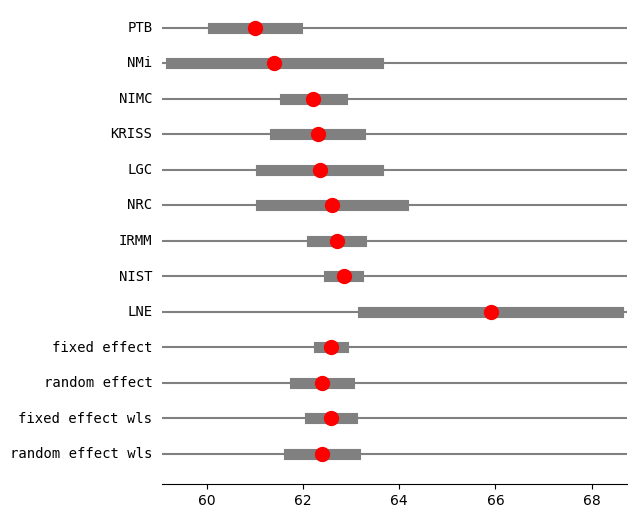
res2_DL = combine_effects(eff, var_eff, method_re="dl", use_t=True, row_names=rownames)
print("method RE:", res2_DL.method_re)
print(res2_DL.summary_frame())
fig = res2_DL.plot_forest()
fig.set_figheight(6)
fig.set_figwidth(6)
method RE: dl
eff sd_eff ci_low ci_upp w_fe w_re
PTB 61.000000 0.450000 60.118016 61.881984 0.057436 0.123113
NMi 61.400000 1.100000 59.244040 63.555960 0.009612 0.040314
NIMC 62.210000 0.300000 61.622011 62.797989 0.129230 0.159749
KRISS 62.300000 0.450000 61.418016 63.181984 0.057436 0.123113
LGC 62.340000 0.620000 61.124822 63.555178 0.030257 0.089810
NRC 62.600000 0.750000 61.130027 64.069973 0.020677 0.071005
IRMM 62.700000 0.260000 62.190409 63.209591 0.172052 0.169810
NIST 62.840000 0.150000 62.546005 63.133995 0.516920 0.194471
LNE 65.900000 1.350000 63.254049 68.545951 0.006382 0.028615
fixed effect 62.583397 0.107846 62.334704 62.832090 1.000000 NaN
random effect 62.390139 0.245750 61.823439 62.956838 NaN 1.000000
fixed effect wls 62.583397 0.189889 62.145512 63.021282 1.000000 NaN
random effect wls 62.390139 0.294776 61.710384 63.069893 NaN 1.000000
/opt/hostedtoolcache/Python/3.10.15/x64/lib/python3.10/site-packages/statsmodels/stats/meta_analysis.py:105: UserWarning: `use_t=True` requires `nobs` for each sample or `ci_func`. Using normal distribution for confidence interval of individual samples.
warnings.warn(msg)
[15]:
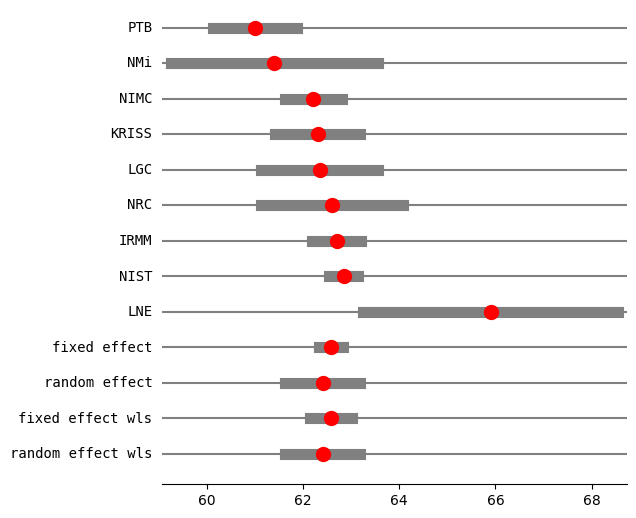
res2_PM = combine_effects(eff, var_eff, method_re="pm", use_t=True, row_names=rownames)
print("method RE:", res2_PM.method_re)
print(res2_PM.summary_frame())
fig = res2_PM.plot_forest()
fig.set_figheight(6)
fig.set_figwidth(6)
method RE: pm
eff sd_eff ci_low ci_upp w_fe w_re
PTB 61.000000 0.450000 60.118016 61.881984 0.057436 0.125857
NMi 61.400000 1.100000 59.244040 63.555960 0.009612 0.059656
NIMC 62.210000 0.300000 61.622011 62.797989 0.129230 0.143658
KRISS 62.300000 0.450000 61.418016 63.181984 0.057436 0.125857
LGC 62.340000 0.620000 61.124822 63.555178 0.030257 0.104850
NRC 62.600000 0.750000 61.130027 64.069973 0.020677 0.090122
IRMM 62.700000 0.260000 62.190409 63.209591 0.172052 0.147821
NIST 62.840000 0.150000 62.546005 63.133995 0.516920 0.156980
LNE 65.900000 1.350000 63.254049 68.545951 0.006382 0.045201
fixed effect 62.583397 0.107846 62.334704 62.832090 1.000000 NaN
random effect 62.407620 0.338030 61.628120 63.187119 NaN 1.000000
fixed effect wls 62.583397 0.189889 62.145512 63.021282 1.000000 NaN
random effect wls 62.407620 0.338030 61.628120 63.187120 NaN 1.000000
/opt/hostedtoolcache/Python/3.10.15/x64/lib/python3.10/site-packages/statsmodels/stats/meta_analysis.py:105: UserWarning: `use_t=True` requires `nobs` for each sample or `ci_func`. Using normal distribution for confidence interval of individual samples.
warnings.warn(msg)
[ ]:
比例的元分析¶
在下面的示例中,随机效应方差 tau 估计为零。然后我更改了数据中的两个计数,因此第二个示例的随机效应方差大于零。
[16]:
import io
[17]:
ss = """\
study,nei,nci,e1i,c1i,e2i,c2i,e3i,c3i,e4i,c4i
1,19,22,16.0,20.0,11,12,4.0,8.0,4,3
2,34,35,22.0,22.0,18,12,15.0,8.0,15,6
3,72,68,44.0,40.0,21,15,10.0,3.0,3,0
4,22,20,19.0,12.0,14,5,5.0,4.0,2,3
5,70,32,62.0,27.0,42,13,26.0,6.0,15,5
6,183,94,130.0,65.0,80,33,47.0,14.0,30,11
7,26,50,24.0,30.0,13,18,5.0,10.0,3,9
8,61,55,51.0,44.0,37,30,19.0,19.0,11,15
9,36,25,30.0,17.0,23,12,13.0,4.0,10,4
10,45,35,43.0,35.0,19,14,8.0,4.0,6,0
11,246,208,169.0,139.0,106,76,67.0,42.0,51,35
12,386,141,279.0,97.0,170,46,97.0,21.0,73,8
13,59,32,56.0,30.0,34,17,21.0,9.0,20,7
14,45,15,42.0,10.0,18,3,9.0,1.0,9,1
15,14,18,14.0,18.0,13,14,12.0,13.0,9,12
16,26,19,21.0,15.0,12,10,6.0,4.0,5,1
17,74,75,,,42,40,,,23,30"""
df3 = pd.read_csv(io.StringIO(ss))
df_12y = df3[["e2i", "nei", "c2i", "nci"]]
# TODO: currently 1 is reference, switch labels
count1, nobs1, count2, nobs2 = df_12y.values.T
dta = df_12y.values.T
[18]:
eff, var_eff = effectsize_2proportions(*dta, statistic="rd")
[19]:
eff, var_eff
[19]:
(array([ 0.03349282, 0.18655462, 0.07107843, 0.38636364, 0.19375 ,
0.08609464, 0.14 , 0.06110283, 0.15888889, 0.02222222,
0.06550969, 0.11417337, 0.04502119, 0.2 , 0.15079365,
-0.06477733, 0.03423423]),
array([0.02409958, 0.01376482, 0.00539777, 0.01989341, 0.01096641,
0.00376814, 0.01422338, 0.00842011, 0.01639261, 0.01227827,
0.00211165, 0.00219739, 0.01192067, 0.016 , 0.0143398 ,
0.02267994, 0.0066352 ]))
[20]:
res5 = combine_effects(
eff, var_eff, method_re="iterated", use_t=False
) # , row_names=rownames)
res5_df = res5.summary_frame()
print("method RE:", res5.method_re)
print("RE variance tau2:", res5.tau2)
print(res5.summary_frame())
fig = res5.plot_forest()
fig.set_figheight(8)
fig.set_figwidth(6)
method RE: iterated
RE variance tau2: 0
eff sd_eff ci_low ci_upp w_fe w_re
0 0.033493 0.155240 -0.270773 0.337758 0.017454 0.017454
1 0.186555 0.117324 -0.043395 0.416505 0.030559 0.030559
2 0.071078 0.073470 -0.072919 0.215076 0.077928 0.077928
3 0.386364 0.141044 0.109922 0.662805 0.021145 0.021145
4 0.193750 0.104721 -0.011499 0.398999 0.038357 0.038357
5 0.086095 0.061385 -0.034218 0.206407 0.111630 0.111630
6 0.140000 0.119262 -0.093749 0.373749 0.029574 0.029574
7 0.061103 0.091761 -0.118746 0.240951 0.049956 0.049956
8 0.158889 0.128034 -0.092052 0.409830 0.025660 0.025660
9 0.022222 0.110807 -0.194956 0.239401 0.034259 0.034259
10 0.065510 0.045953 -0.024556 0.155575 0.199199 0.199199
11 0.114173 0.046876 0.022297 0.206049 0.191426 0.191426
12 0.045021 0.109182 -0.168971 0.259014 0.035286 0.035286
13 0.200000 0.126491 -0.047918 0.447918 0.026290 0.026290
14 0.150794 0.119749 -0.083910 0.385497 0.029334 0.029334
15 -0.064777 0.150599 -0.359945 0.230390 0.018547 0.018547
16 0.034234 0.081457 -0.125418 0.193887 0.063395 0.063395
fixed effect 0.096212 0.020509 0.056014 0.136410 1.000000 NaN
random effect 0.096212 0.020509 0.056014 0.136410 NaN 1.000000
fixed effect wls 0.096212 0.016521 0.063831 0.128593 1.000000 NaN
random effect wls 0.096212 0.016521 0.063831 0.128593 NaN 1.000000
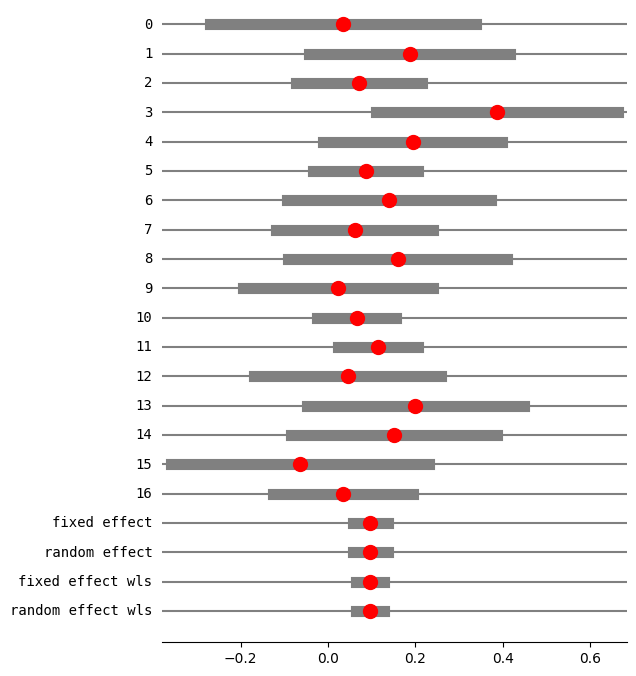
更改数据以具有正的随机效应方差¶
[21]:
dta_c = dta.copy()
dta_c.T[0, 0] = 18
dta_c.T[1, 0] = 22
dta_c.T
[21]:
array([[ 18, 19, 12, 22],
[ 22, 34, 12, 35],
[ 21, 72, 15, 68],
[ 14, 22, 5, 20],
[ 42, 70, 13, 32],
[ 80, 183, 33, 94],
[ 13, 26, 18, 50],
[ 37, 61, 30, 55],
[ 23, 36, 12, 25],
[ 19, 45, 14, 35],
[106, 246, 76, 208],
[170, 386, 46, 141],
[ 34, 59, 17, 32],
[ 18, 45, 3, 15],
[ 13, 14, 14, 18],
[ 12, 26, 10, 19],
[ 42, 74, 40, 75]])
[22]:
eff, var_eff = effectsize_2proportions(*dta_c, statistic="rd")
res5 = combine_effects(
eff, var_eff, method_re="iterated", use_t=False
) # , row_names=rownames)
res5_df = res5.summary_frame()
print("method RE:", res5.method_re)
print(res5.summary_frame())
fig = res5.plot_forest()
fig.set_figheight(8)
fig.set_figwidth(6)
method RE: iterated
eff sd_eff ci_low ci_upp w_fe w_re
0 0.401914 0.117873 0.170887 0.632940 0.029850 0.038415
1 0.304202 0.114692 0.079410 0.528993 0.031529 0.040258
2 0.071078 0.073470 -0.072919 0.215076 0.076834 0.081017
3 0.386364 0.141044 0.109922 0.662805 0.020848 0.028013
4 0.193750 0.104721 -0.011499 0.398999 0.037818 0.046915
5 0.086095 0.061385 -0.034218 0.206407 0.110063 0.102907
6 0.140000 0.119262 -0.093749 0.373749 0.029159 0.037647
7 0.061103 0.091761 -0.118746 0.240951 0.049255 0.058097
8 0.158889 0.128034 -0.092052 0.409830 0.025300 0.033270
9 0.022222 0.110807 -0.194956 0.239401 0.033778 0.042683
10 0.065510 0.045953 -0.024556 0.155575 0.196403 0.141871
11 0.114173 0.046876 0.022297 0.206049 0.188739 0.139144
12 0.045021 0.109182 -0.168971 0.259014 0.034791 0.043759
13 0.200000 0.126491 -0.047918 0.447918 0.025921 0.033985
14 0.150794 0.119749 -0.083910 0.385497 0.028922 0.037383
15 -0.064777 0.150599 -0.359945 0.230390 0.018286 0.024884
16 0.034234 0.081457 -0.125418 0.193887 0.062505 0.069751
fixed effect 0.110252 0.020365 0.070337 0.150167 1.000000 NaN
random effect 0.117633 0.024913 0.068804 0.166463 NaN 1.000000
fixed effect wls 0.110252 0.022289 0.066567 0.153937 1.000000 NaN
random effect wls 0.117633 0.024913 0.068804 0.166463 NaN 1.000000
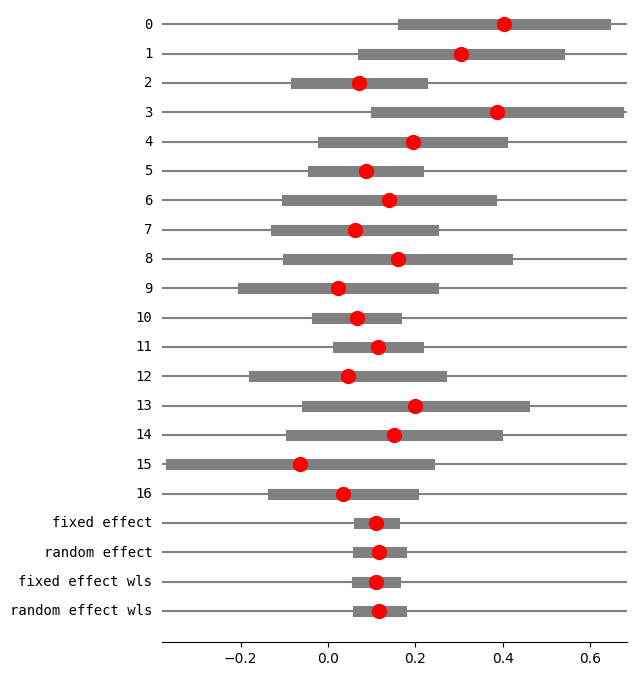
[23]:
res5 = combine_effects(eff, var_eff, method_re="chi2", use_t=False)
res5_df = res5.summary_frame()
print("method RE:", res5.method_re)
print(res5.summary_frame())
fig = res5.plot_forest()
fig.set_figheight(8)
fig.set_figwidth(6)
method RE: chi2
eff sd_eff ci_low ci_upp w_fe w_re
0 0.401914 0.117873 0.170887 0.632940 0.029850 0.036114
1 0.304202 0.114692 0.079410 0.528993 0.031529 0.037940
2 0.071078 0.073470 -0.072919 0.215076 0.076834 0.080779
3 0.386364 0.141044 0.109922 0.662805 0.020848 0.025973
4 0.193750 0.104721 -0.011499 0.398999 0.037818 0.044614
5 0.086095 0.061385 -0.034218 0.206407 0.110063 0.105901
6 0.140000 0.119262 -0.093749 0.373749 0.029159 0.035356
7 0.061103 0.091761 -0.118746 0.240951 0.049255 0.056098
8 0.158889 0.128034 -0.092052 0.409830 0.025300 0.031063
9 0.022222 0.110807 -0.194956 0.239401 0.033778 0.040357
10 0.065510 0.045953 -0.024556 0.155575 0.196403 0.154854
11 0.114173 0.046876 0.022297 0.206049 0.188739 0.151236
12 0.045021 0.109182 -0.168971 0.259014 0.034791 0.041435
13 0.200000 0.126491 -0.047918 0.447918 0.025921 0.031761
14 0.150794 0.119749 -0.083910 0.385497 0.028922 0.035095
15 -0.064777 0.150599 -0.359945 0.230390 0.018286 0.022976
16 0.034234 0.081457 -0.125418 0.193887 0.062505 0.068449
fixed effect 0.110252 0.020365 0.070337 0.150167 1.000000 NaN
random effect 0.115580 0.023557 0.069410 0.161751 NaN 1.000000
fixed effect wls 0.110252 0.022289 0.066567 0.153937 1.000000 NaN
random effect wls 0.115580 0.024241 0.068068 0.163093 NaN 1.000000
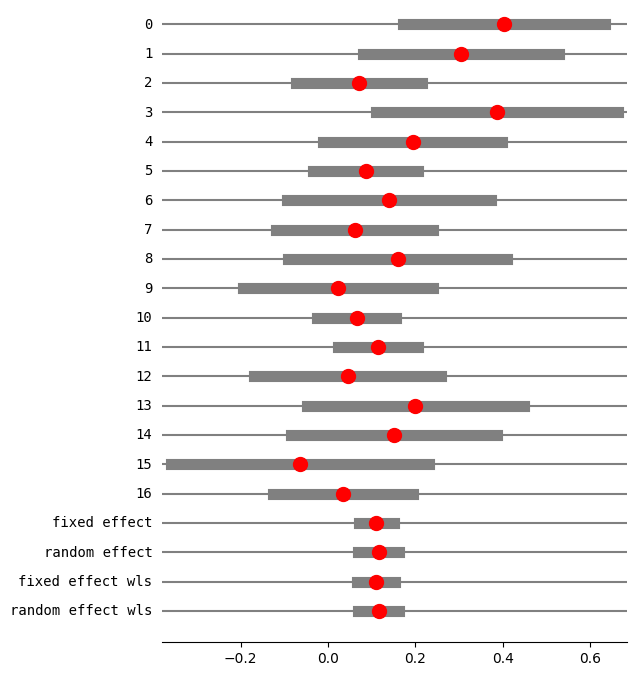
使用 GLM 和 var_weights 复制固定效应分析¶
combine_effects 计算加权平均估计,可以使用 GLM 和 var_weights 或 WLS 复制。在 GLM.fit 中的 scale 选项可用于使用固定比例和 HKSJ/WLS 比例复制固定元分析
[24]:
from statsmodels.genmod.generalized_linear_model import GLM
[25]:
eff, var_eff = effectsize_2proportions(*dta_c, statistic="or")
res = combine_effects(eff, var_eff, method_re="chi2", use_t=False)
res_frame = res.summary_frame()
print(res_frame.iloc[-4:])
eff sd_eff ci_low ci_upp w_fe w_re
fixed effect 0.428037 0.090287 0.251076 0.604997 1.0 NaN
random effect 0.429520 0.091377 0.250425 0.608615 NaN 1.0
fixed effect wls 0.428037 0.090798 0.250076 0.605997 1.0 NaN
random effect wls 0.429520 0.091595 0.249997 0.609044 NaN 1.0
我们需要将 scale 固定为 1,以复制通常的元分析的标准误。
[26]:
weights = 1 / var_eff
mod_glm = GLM(eff, np.ones(len(eff)), var_weights=weights)
res_glm = mod_glm.fit(scale=1.0)
print(res_glm.summary().tables[1])
==============================================================================
coef std err z P>|z| [0.025 0.975]
------------------------------------------------------------------------------
const 0.4280 0.090 4.741 0.000 0.251 0.605
==============================================================================
[27]:
# check results
res_glm.scale, res_glm.conf_int() - res_frame.loc[
"fixed effect", ["ci_low", "ci_upp"]
].values
[27]:
(array(1.), array([[-5.55111512e-17, 0.00000000e+00]]))
在元分析中使用 HKSJ 方差调整等效于使用 pearson chi2 估计比例,这也是高斯族的默认值。
[28]:
res_glm = mod_glm.fit(scale="x2")
print(res_glm.summary().tables[1])
==============================================================================
coef std err z P>|z| [0.025 0.975]
------------------------------------------------------------------------------
const 0.4280 0.091 4.714 0.000 0.250 0.606
==============================================================================
[29]:
# check results
res_glm.scale, res_glm.conf_int() - res_frame.loc[
"fixed effect", ["ci_low", "ci_upp"]
].values
[29]:
(np.float64(1.0113358914264383), array([[-0.00100017, 0.00100017]]))
使用列联表进行 Mantel-Hanszel 优势比¶
可以使用 StratifiedTable 直接计算使用 Mantel-Hanszel 的对数优势比的固定效应。
我们需要创建一个 2 x 2 x k 列联表,用于与 StratifiedTable 一起使用。
[30]:
t, nt, c, nc = dta_c
counts = np.column_stack([t, nt - t, c, nc - c])
ctables = counts.T.reshape(2, 2, -1)
ctables[:, :, 0]
[30]:
array([[18, 1],
[12, 10]])
[31]:
counts[0]
[31]:
array([18, 1, 12, 10])
[32]:
dta_c.T[0]
[32]:
array([18, 19, 12, 22])
[33]:
import statsmodels.stats.api as smstats
[34]:
st = smstats.StratifiedTable(ctables.astype(np.float64))
将合并的对数优势比和标准误与 R meta 包进行比较
[35]:
st.logodds_pooled, st.logodds_pooled - 0.4428186730553189 # R meta
[35]:
(np.float64(0.4428186730553187), np.float64(-2.220446049250313e-16))
[36]:
st.logodds_pooled_se, st.logodds_pooled_se - 0.08928560091027186 # R meta
[36]:
(np.float64(0.08928560091027186), np.float64(0.0))
[37]:
st.logodds_pooled_confint()
[37]:
(np.float64(0.2678221109331691), np.float64(0.6178152351774683))
[38]:
print(st.test_equal_odds())
pvalue 0.34496419319878724
statistic 17.64707987033203
[39]:
print(st.test_null_odds())
pvalue 6.615053645964153e-07
statistic 24.724136624311814
检查转换为分层列联表
每个表的行总和是治疗组和对照组实验的样本量
[40]:
ctables.sum(1)
[40]:
array([[ 19, 34, 72, 22, 70, 183, 26, 61, 36, 45, 246, 386, 59,
45, 14, 26, 74],
[ 22, 35, 68, 20, 32, 94, 50, 55, 25, 35, 208, 141, 32,
15, 18, 19, 75]])
[41]:
nt, nc
[41]:
(array([ 19, 34, 72, 22, 70, 183, 26, 61, 36, 45, 246, 386, 59,
45, 14, 26, 74]),
array([ 22, 35, 68, 20, 32, 94, 50, 55, 25, 35, 208, 141, 32,
15, 18, 19, 75]))
R meta 包的结果
> res_mb_hk = metabin(e2i, nei, c2i, nci, data=dat2, sm="OR", Q.Cochrane=FALSE, method="MH", method.tau="DL", hakn=FALSE, backtransf=FALSE)
> res_mb_hk
logOR 95%-CI %W(fixed) %W(random)
1 2.7081 [ 0.5265; 4.8896] 0.3 0.7
2 1.2567 [ 0.2658; 2.2476] 2.1 3.2
3 0.3749 [-0.3911; 1.1410] 5.4 5.4
4 1.6582 [ 0.3245; 2.9920] 0.9 1.8
5 0.7850 [-0.0673; 1.6372] 3.5 4.4
6 0.3617 [-0.1528; 0.8762] 12.1 11.8
7 0.5754 [-0.3861; 1.5368] 3.0 3.4
8 0.2505 [-0.4881; 0.9892] 6.1 5.8
9 0.6506 [-0.3877; 1.6889] 2.5 3.0
10 0.0918 [-0.8067; 0.9903] 4.5 3.9
11 0.2739 [-0.1047; 0.6525] 23.1 21.4
12 0.4858 [ 0.0804; 0.8911] 18.6 18.8
13 0.1823 [-0.6830; 1.0476] 4.6 4.2
14 0.9808 [-0.4178; 2.3795] 1.3 1.6
15 1.3122 [-1.0055; 3.6299] 0.4 0.6
16 -0.2595 [-1.4450; 0.9260] 3.1 2.3
17 0.1384 [-0.5076; 0.7844] 8.5 7.6
Number of studies combined: k = 17
logOR 95%-CI z p-value
Fixed effect model 0.4428 [0.2678; 0.6178] 4.96 < 0.0001
Random effects model 0.4295 [0.2504; 0.6086] 4.70 < 0.0001
Quantifying heterogeneity:
tau^2 = 0.0017 [0.0000; 0.4589]; tau = 0.0410 [0.0000; 0.6774];
I^2 = 1.1% [0.0%; 51.6%]; H = 1.01 [1.00; 1.44]
Test of heterogeneity:
Q d.f. p-value
16.18 16 0.4404
Details on meta-analytical method:
- Mantel-Haenszel method
- DerSimonian-Laird estimator for tau^2
- Jackson method for confidence interval of tau^2 and tau
> res_mb_hk$TE.fixed
[1] 0.4428186730553189
> res_mb_hk$seTE.fixed
[1] 0.08928560091027186
> c(res_mb_hk$lower.fixed, res_mb_hk$upper.fixed)
[1] 0.2678221109331694 0.6178152351774684
[42]:
print(st.summary())
Estimate LCB UCB
-----------------------------------------
Pooled odds 1.557 1.307 1.855
Pooled log odds 0.443 0.268 0.618
Pooled risk ratio 1.270
Statistic P-value
-----------------------------------
Test of OR=1 24.724 0.000
Test constant OR 17.647 0.345
-----------------------
Number of tables 17
Min n 32
Max n 527
Avg n 139
Total n 2362
-----------------------